Metabolomic investigation of polyketide biosynthesis in Streptomyces cinnamonensis скачать в хорошем качестве
Повторяем попытку...
Скачать видео с ютуб по ссылке или смотреть без блокировок на сайте: Metabolomic investigation of polyketide biosynthesis in Streptomyces cinnamonensis в качестве 4k
У нас вы можете посмотреть бесплатно Metabolomic investigation of polyketide biosynthesis in Streptomyces cinnamonensis или скачать в максимальном доступном качестве, видео которое было загружено на ютуб. Для загрузки выберите вариант из формы ниже:
-
Информация по загрузке:
Скачать mp3 с ютуба отдельным файлом. Бесплатный рингтон Metabolomic investigation of polyketide biosynthesis in Streptomyces cinnamonensis в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием видео, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса ClipSaver.ru
Metabolomic investigation of polyketide biosynthesis in Streptomyces cinnamonensis
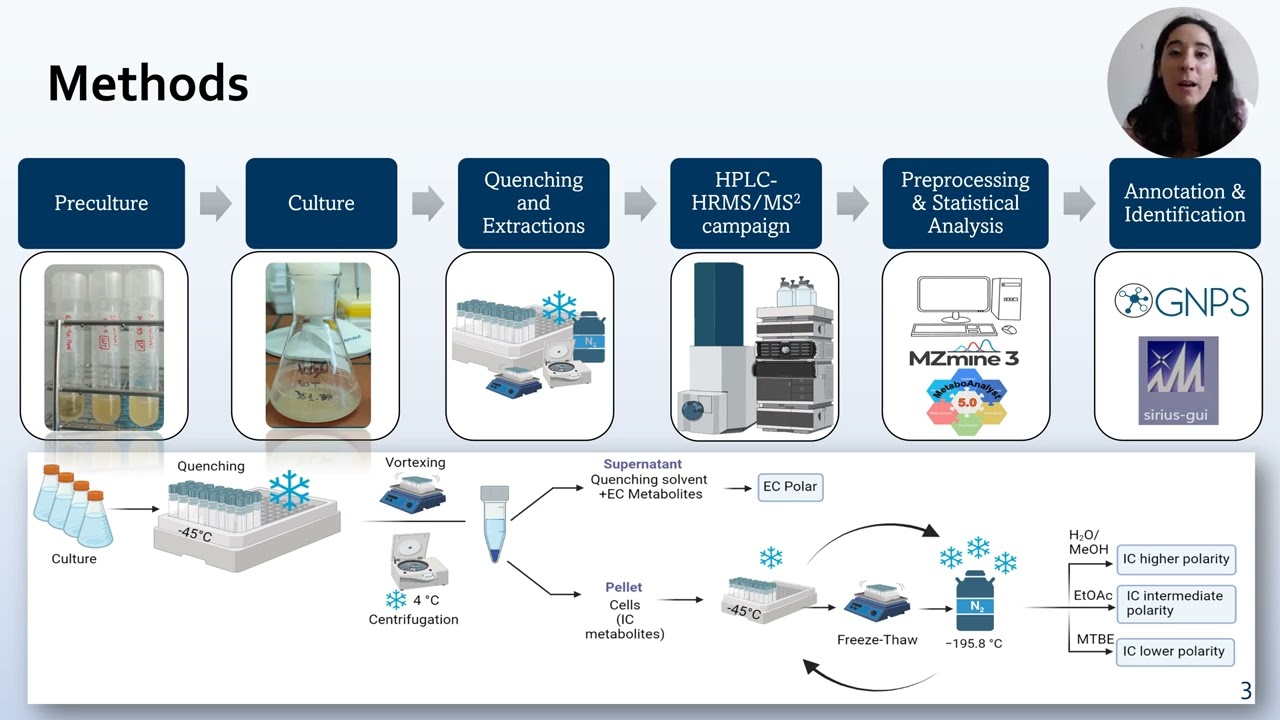
The genus Streptomyces is a known rich source of bioactive compounds [1]. Streptomyces cinnamonensis produces the antibiotic monensin, which is synthesized by a multi-enzyme complex from the Polyketide Synthase family. This species is of interest due to the possibility of predictably modifying the catalytic domains to biosynthesize a library of novel bioactive compounds. However, the often-low fermentation titers hamper the production of new compounds for biological testing. To increase the production of new-to-nature molecules, enzymatic bottlenecks need to be detected and relieved. These bottlenecks can be reflected by the accumulation of characteristic metabolites along the mutated biosynthetic pathway. This principle can be applied to both the new-to-nature producers and classical monensin overproducing strains. Initially, comparative targeted metabolomics of monensin biosynthetic intermediates was applied to different mutant strains. Polyketide intermediates were extracted from liquid cultures and analyzed on an LC-HRMS system (Bruker QTOF). Next, this was extended to the whole metabolism, aimed at overproducing polyketides. Data were analyzed using DataAnalysis (Bruker Daltonics), Metaboscape (Bruker Daltonics), or MZmine3. The comparative metabolomic investigation led to the identification of biosynthetic bottlenecks toward new-to-nature compounds. Non-natural intermediates accumulated in the biosynthetic flux downstream of the mutagenesis sites and were identified by MS2 and, in part, by NMR. This indicates that the processivity of new-to-nature metabolites is greatly reduced compared to the wildtype. This study expanded to a global view, considering the impact of this biosynthesis on the whole organism. In an untargeted manner, the wild-type S. cinnamonensis and some of its variants were compared to an industrial monensin overproducer to identify the biosynthetic bottlenecks for overproduction. The latest data revealed the relevance of regulatory compounds on several metabolic pathways, and the data are currently being linked to comparative genomics. The knowledge gained from this experiment will be used to increase the production of new-to-nature polyketides. [1] Gavriilidou A, Kautsar SA, Zaburannyi N, Krug D, Müller R, Medema MH, Ziemert N. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nat Microbiol. 2022 7(5):726-735.